Segmentation and Material Identification
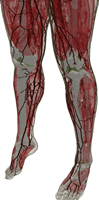
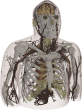
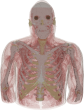
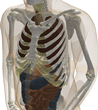
The filtered (masked) image slices were segmented into regions and each region was assigned an electromagnetic material id. The authors completed the segmentation/material identification step in several stages, which resulted in different models. Initially, a 4 mm z-resolution model was developed by (independently) segmenting every fourth slice 1002, 1006, 1010, etc. The segmentation was accomplished first by using a region growing algorithm that grouped pixels similar in color [8]. Then manual adjustments were made to region boundaries using the Boundary and Region Identification Software Kit (BRISKit) developed in MATLAB [8]. The correctness of the boundaries is limited by the image resolution. In color images, this is manifested by ill-defined transition regions, where the colors are mixed and pixels do not clearly belong to one region/material; e.g., in this figure of slice 1070, zooming in to the region outlined by the blue box in the middle image shows the pixilated transition between the white matter and grey matter in the brain on the left and the resulting outline of the grey matter on the right (where the green boxes represent the edge of the grey matter, inclusively). During the segmentation process in the AustinMan model, material boundaries were identified such that they are accurate to within 3 pixels (±1 mm) of the actual boundary. There are some exceptions where the boundaries are particularly difficult to identify and thus less precise; these are listed in the Problem Regions section.
Once the region boundaries were finalized in each masked image slice, anatomical parts were identified with the help of the Atlas of the Visible Human Male [9] and an electromagnetic material id was assigned to each region. The material ids are the same as the 56 materials listed in [10]. Not all parts in the human anatomy have a corresponding material with known electromagnetic properties, e.g., the ventricles of the brain. The authors have used their anatomy knowledge and best judgment to assign them to (the most similar) one of the 56 materials in [9], e.g., the ventricles of the brain were assigned to cerebrospinal fluid. These material assignments are listed in detail in the Tissue Material Assumptions section.
After the 4 mm z-resolution model was completed, an interpolation-based approach was adopted to reduce the amount of manual labor. Once the segmentation/material identification step was completed for an image slice, these were used as the initial guess for the slice immediately above or below that slice. The boundaries were then interactively adjusted by using a local-competition algorithm that compared the colors of pixels at the boundaries of the regions to the average color of a set of pixels (manually selected to represent different regions) [8]. The boundary pixels were then reassigned to the region that they are closest to in color (again using the L*a*b* color space) and the process was iterated until the boundary pixels no longer changed or satisfactorily represented the material boundary in the picture. In this interpolation-based approach, manual adjustments were still needed to improve the accuracy of the boundary and material identification; however, less time had to be spent on each slice.
| Previous: Masking | Up: Methodology | Next: Coarsening and Extrusion |