![[OK]](Icons/tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Sample_Yeast_L005_R1.cat.fastq.gz |
| File type | Conventional base calls |
| Encoding | Sanger / Illumina 1.9 |
| Total Sequences | 592180 |
| Filtered Sequences | 0 |
| Sequence length | 101 |
| %GC | 40 |
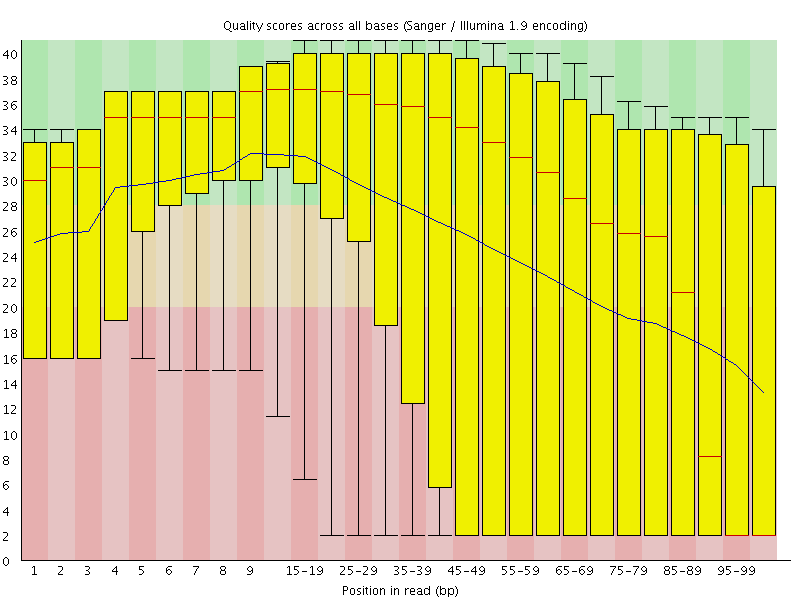
![[FAIL]](Icons/error.png) Per base sequence quality
Per base sequence quality
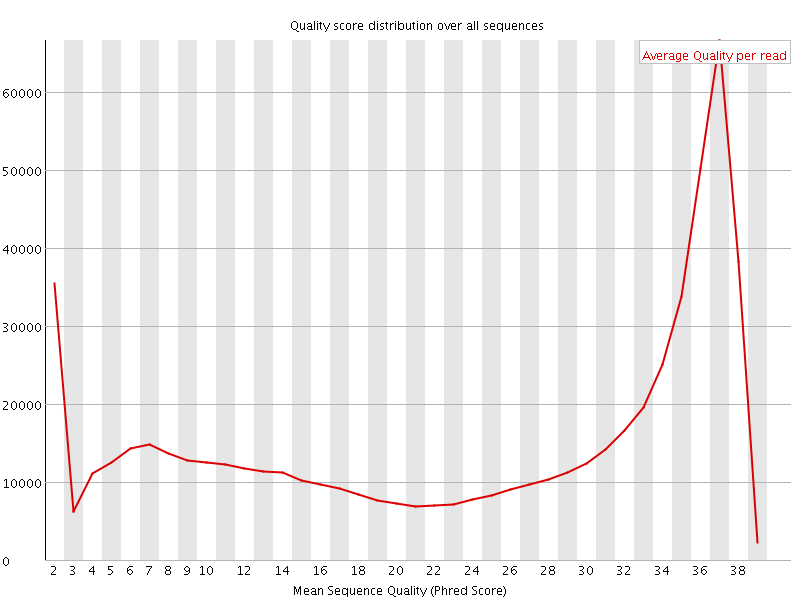
![[OK]](Icons/tick.png) Per sequence quality scores
Per sequence quality scores
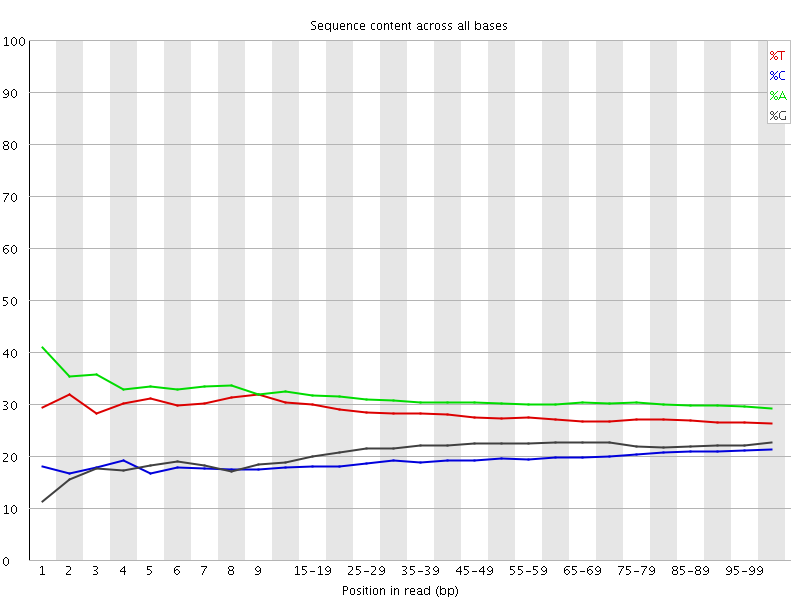
![[FAIL]](Icons/error.png) Per base sequence content
Per base sequence content
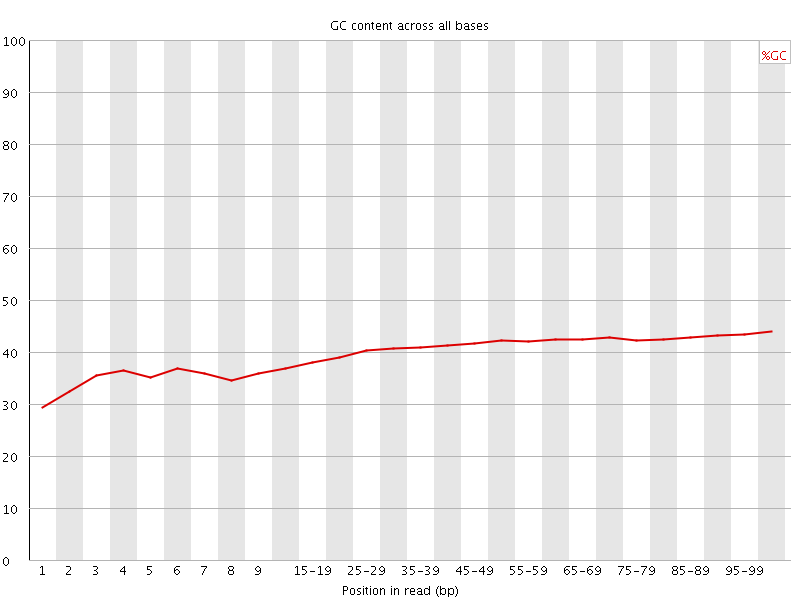
![[FAIL]](Icons/error.png) Per base GC content
Per base GC content
![[WARN]](Icons/warning.png) Per sequence GC content
Per sequence GC content
![[OK]](Icons/tick.png) Per base N content
Per base N content
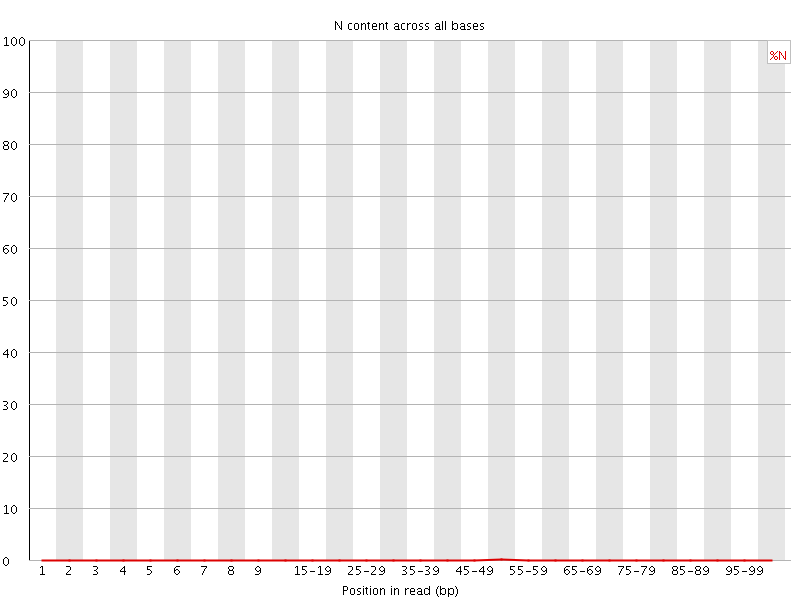
![[OK]](Icons/tick.png) Sequence Length Distribution
Sequence Length Distribution
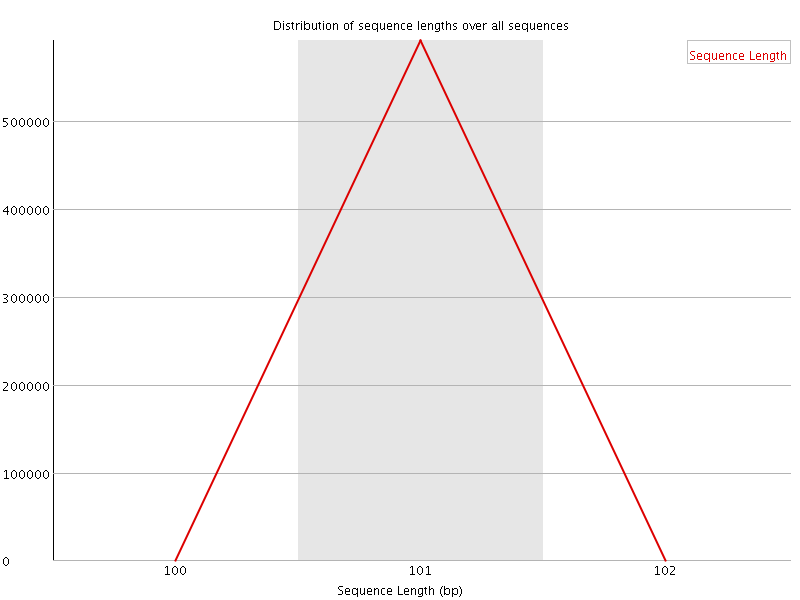
![[WARN]](Icons/warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[WARN]](Icons/warning.png) Overrepresented sequences
Overrepresented sequences
| Sequence | Count | Percentage | Possible Source |
|---|---|---|---|
| GATCGGAAGAGCACACGTCTGAACTCCAGTCACCTCAGAATCTCGTATGC | 2803 | 0.4733358100577527 | TruSeq Adapter, Index 1 (97% over 37bp) |
![[WARN]](Icons/warning.png) Kmer Content
Kmer Content
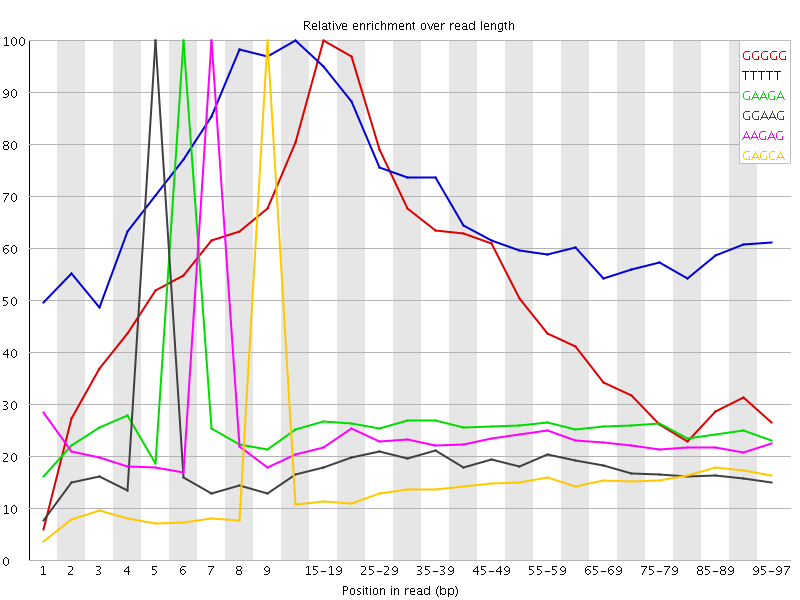
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGGGG | 110810 | 4.19239 | 7.96175 | 15-19 |
| TTTTT | 352785 | 3.5414593 | 5.207852 | 10-14 |
| GAAGA | 138530 | 1.7625622 | 6.7152853 | 6 |
| GGAAG | 87000 | 1.5917709 | 8.4951315 | 5 |
| AAGAG | 113955 | 1.4498866 | 6.222126 | 7 |
| GAGCA | 63940 | 1.2896557 | 8.680826 | 9 |
| AGAGC | 63350 | 1.2777555 | 8.778584 | 8 |
| CGGAA | 47700 | 0.96209836 | 8.886041 | 4 |
| ATCGG | 33030 | 0.7347307 | 9.608717 | 2 |
| TCGGA | 32035 | 0.7125976 | 9.315497 | 3 |
| GATCG | 28845 | 0.64163816 | 8.56018 | 1 |